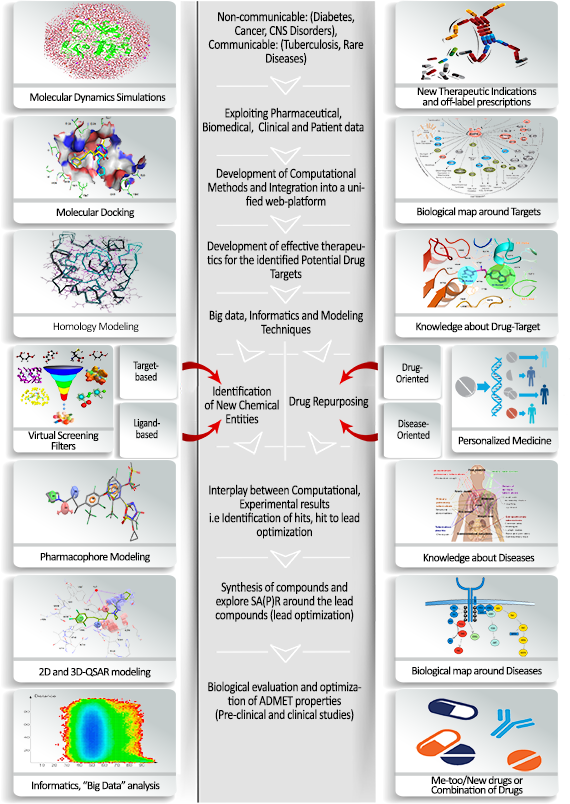
The Project
The purpose of this project is to develop a computational platform designed for efficient analysis and interpretation of the extensive biomedical and clinical data, the comparative analysis extracted from these data information to differentiate the normal and pathological states, obtaining new knowledge to identify biomarkers and pharmacological targets, designing potential drugs with the required properties, determine the optimal approach to therapy taking into account the individuality of patients. The project will be carried out by analyzing existing data sources and selecting the most promising information and computational resources; if necessary, available methods will be improved, and new methods will be created; an approach to their integration into a single platform will be developed and implemented (see: General Flowchart).
Validation of the developed computational platform will be carried out on the case studies of common and rare socially important diseases with non-infectious nature (diabetes, cancer, CNS disorders) and infectious diseases (tuberculosis, leishmaniasis, schistosomiasis), which will determine the optimal scenario for its use and, if necessary, to conduct an appropriate upgrade.
Special emphasis will be placed on the repositioning of known drugs (identifying new indications for known pharmaceuticals), which allows in many cases to carry out an accelerated testing of computer-aided predictions directly in the clinic and even a direct introduction of such drugs into medical practice ("off-label prescription"). Informatics based approaches with the data available from the physicians will be employed for the identification of optimal FDA approved drugs for their new pharmacological effects.
General Flowchart
Collaboration
Center for Molecular Modeling of the CSIR-Institute of Chemical Technology headed by Prof. Narahari Sastry for a number of years engaged in the development and practical application of computer-aided drug design methods, including structure-based approaches, pharmacophore modeling, virtual screening, etc. They designed new pharmacological agents acting on different pharmacological targets, using a wide range of molecular modeling techniques, such as molecular dynamics, docking, homology modeling, QSAR and informatics methods. Indian team created the Molecular Property Diagnostic Suite (MPDS) on the basis of Galaxy platform. The main aim of MPDS is to assess and estimate the drug-likeliness of a particular compound in order to estimate its potential applicability as a drug. MPDS is a common platform to merge many available open-source as well as in-house in silico tools to support open source drug discovery programs by prioritizing compounds before taking to synthesis.
Indian team made a number of predictions about the properties of synthetic accessibility and properties of organic molecules. A large part of computational predictions was confirmed experimentally (PLoS ONE., 2015, 10: e0144294, J. Phys. Chem. C., 2015, 119: 23607–23618, J. Chem. Inf. Model., 2015, 55: 848-860, J. Comp. Chem., 2015, 36: 529-538, PLoSONE, 2014, 9: e113773, Acc. Chem. Res., 2014, 47: 2574-2581, J. Comp. Chem., 2014, 35: 595-610, Curr. Pharm. Des., 2013, 19: 4714-4738, Chem. Rev., 2013, 113: 2100-2138, J. Chem. Inf. Model., 2011, 51: 558-571).
Laboratory for Structure-Function Based Drug Design headed by Prof. Vladimir Poroikov during the past twenty years develops computational methods, programs and databases for identification of promising pharmacological targets and finding their ligands. An original system of atom-centric molecular descriptors characterized the intermolecular protein-ligand interactions is developed, and used as the basis to perform analysis of various structure-property relationships. (J. Chem. Inf. Comput. Sci., 1999, 39: 666–670; SAR & QSAR Environ. Res., 2009, 20: 679-709; J. Chem. Inf. Model., 2014, 54: 498–507). Russian team created computer programs for prediction on the basis of structural formula of drug-like organic compounds: biological activity spectra (Chem. Heterocycl. Comp., 2014, 50: 444-457); gene expression (Bioinformatics, 2013, 29: 2062-2063); acute toxicity to rats at four routes of administration (Mol. Inform., 2011, 30: 241–250); interaction with undesired targets (Chem. Res. Toxicol., 2012, 25: 2378-2385), interaction with biological targets (SAR & QSAR Environ. Res., 2015, 26: 783-793); interaction with non-tumor and tumor cell lines (SAR & QSAR Environ. Res., 2015, 26: 595-604); interaction with metabolic enzymes (J. Chemoinform., 2016, 8: 68).
Based on the created computational tools, Russian team developed a number of predictive web-services at the Way2Drug platform that are used by many researchers worldwide. The synergistic association of mutually complementary knowledge, experience and skills of multidisciplinary teams of Russia and India in the framework of this project will allow achieving the results that none of the teams could be achieve working separately.
The project will lead to overcome the interdisciplinary barriers between the biomedical and clinical disciplines by creating dynamic data-processing environment for the interaction of diverse specialists that will lead to a significant increase in the efficiency of the use of the extensive biomedical and clinical data in medical practice, and lay the foundation for long-term cooperation between Indian and Russian scientists in the area of discovery, development and application of novel more effective and safe medicines.
Acknowledgements
The project is supported by the Russian Science Foundation (grant No. 16-45-02012) and Indian Department of Science and Technology (grant No. INT/RUS/RSF/12).
Principal Investigators

Prof. Dr. Vladimir Poroikov, Laboratory for Structure-Function Drug Design, Institute of Biomedical Chemistry, Moscow, Russia. CV

Dr. G. Narahari Sastry, Molecular Modeling Group, CSIR-Indian Institute of Chemical Technology, Hyderabad, India. CV
Meetings
On October 30 – November 6, 2018, three Russian participants of the project, Drs. Vladimir Poroikov, Alexey Lagunin and Dmitry Druzhilovskiy, have visited the Punjabi University, Patiala, India. During this visit, they met with the Indian collaborators of the project and discussed the current state and the further work that must be realized by the end of the project. Russian and Indian participants of the project presented the achieved results at the International Conference & Workshop «Informatics Tools in Drug Discovery and Drug Delivery». They also took part in the International Symposium "Informatics Tools in Drug Discovery and Delivery" that was organized by Chitkara University College of Pharmacy jointly with the Association of Pharmaceutical Teachers of India (APTI), Punjab.
On August 6-12, 2018, Dr. Rajesh Kumar Goel has visited the Russian collaborators at the Institute of Biomedical Chemistry, Moscow, Russia. During this meeting, the participants of the project discussed the current state of the project and the further work that must be realized in 2018.
On October 21-26, 2017, two members of Russian team, Drs. Vladimir Poroikov and Dmitry Druzhilovskiy, have visited CSIR-Institute of Chemical Technology (CSIR-IICT) in Hyderabad, India. During this meeting, Russian and Indian participants of the project discussed the status of the project, reviewed the work that was implemented in the year 2016-2017 and planned for the work to be carried out for the year 2017-2018. On October 23, 2017 one day National Workshop "A Big Data Applications in Drug Repurposing (BDADR-17)" was organized at the GITAM University. At the Workshop Prof. Vladimir Poroikov presented a keynote lecture, and some other Indian and Russian participants of our joint project presented four oral talks.
Then, both Russian and Indian participants of the project presented the achieved results at the 9th International Symposium on Computational Methods in Toxicology and Pharmacology Integrating Internet Resources (CMTPI-2017), which has been held in Bogmallo Beach Resort, Goa, India on October 27-30, 2017. Proceedings of the CMTPI-2017 Symposium were published in SAR and QSAR in Environmental Research (volume 28, issues 10 and 11 and volume 29, issues 1 and 2).
On October 30, 2017, Profs. Vladimir Poroikov and Alexey Lagunin also took part in the India-Russia Conference on Repositioning of Drugs / Natural Products for Healthcare as the satellite event of the Symposium.
On June 15-22, 2017, five members of Indian team, including Drs. Garikapati Narahari Sastry, Rajesh Kumar Goel, Rakesh Sahay, Manisha Sahay and Mr. Sandeep Kumar, have visited the Russian collaborators at the Institute of Biomedical Chemistry, Moscow, Russia. During this meeting, Russian and Indian participants of the project discussed the current state of the project and the further work that should be realized in 2017.
On October 29th – November 4th, 2016 three participants of Russian team, Prof. Vladimir Poroikov, Dr. Anastassia Rudik and Dr. Dmitry Druzhilovskiy, have visited the Indian collaborators at the CSIR-Institute of Chemical Technology (CSIR-IICT) in Hyderabad, India. During this visit Russian and Indian participants of the project discussed the general architecture of the computational platform and protocols of the informational exchange, and also performed some joint computational experiments to validate the computational facilities of Indian and Russian teams.
In joint computational experiments we evaluated the possibility of application of molecular modeling methods by their retrospective validation on a specially prepared test sample, comprising 18 structural formulas of novel anti-TB compounds. Using PASS program, Russian team showed that the existing version of the PASS successfully predicts antituberculosic activity for 11 of the 18 compounds. Molecular docking conducted by the Indian colleagues showed that 17 of 18 compounds of the appraised value of the binding energy function (scoring function) are superior to those of the reference compounds 2,6-pyridinedicarboxylic acid. Thus, the combination of computer predicting methods, based on the structure of the ligand, developed by the Russian participants of the project, with the methods of molecular modeling, prepared by the Indian participants of the project, allows improving the quality of the assessments.
We also discussed with medical doctors from Osmania General Hospital and Osmania Medical College, Prof. Rakesh Kumar Sahay (Department of Endocrinology) and Prof. Manisha Sahay, (Department of Nephrology), the possibilities of utilization of available clinical data on pleiotropic action caused the therapeutic and adverse effects of the approved human medicines.
Prof. Vladimir Poroikov presented a talk «Computer-aided drug repurposing: new uses for old drugs or filling gaps in biomedical knowledge?» at the seminar, and, due to the special request of Indian collaborators, presented a talk «Possibilities for Cooperation between Russia and India in Education and Science» at the meeting with the students and young scientists of the CSIR-IICT.
Also, Russian team has met with Dr. USN Murty, Director, National Institute of Pharmaceutical Education and Research (NIPER) and Dr. Rajendra P. Pawar, Head of the Department of Chemistry, Deogiri College, who are not involved directly in the current RSF-DST project, but expressed their sincere interest to collaboration in the framework of the proposed computational platform.
Our Partners







